Edgington-Mitchell Group

“Our laboratory uses a chemical biology approach to understand the mechanisms by which proteases contribute to normal cellular function and how these mechanisms can go wrong to cause disease. Our overarching goals are to establish proteases as diagnostic or prognostic biomarkers for inflammation and cancer and as novel drug targets for the treatment of these diseases.” – Laura Edgington-Mitchell
Research
Proteases are enzymes that cleave peptide bonds of proteins, a process that underlies many physiological functions that support life. Their most basic functions are to control digestion, degrade unneeded proteins, and promote protein homeostasis. However, they also govern tightly regulated signalling cascades that are important for normal physiological functions, including programmed cell death, blood coagulation, hormone processing, antigen presentation, and cell migration. Dysregulated protease activity is therefore a contributing factor in many diseases, including cancer, inflammation and neurodegenerative disorders.
The Edgington-Mitchell Laboratory uses a multidisciplinary approach to understand the contribution of proteases to health and disease. We are developing novel chemical tools to measure and inhibit the activation of diverse cysteine and serine proteases. We apply these tools in vitro, in mouse models of inflammation and cancer, and in clinical samples derived from patients to dissect protease function and to investigate their utility as biomarkers and therapeutic targets in disease.
Development of activity-based probes to measure protease activity
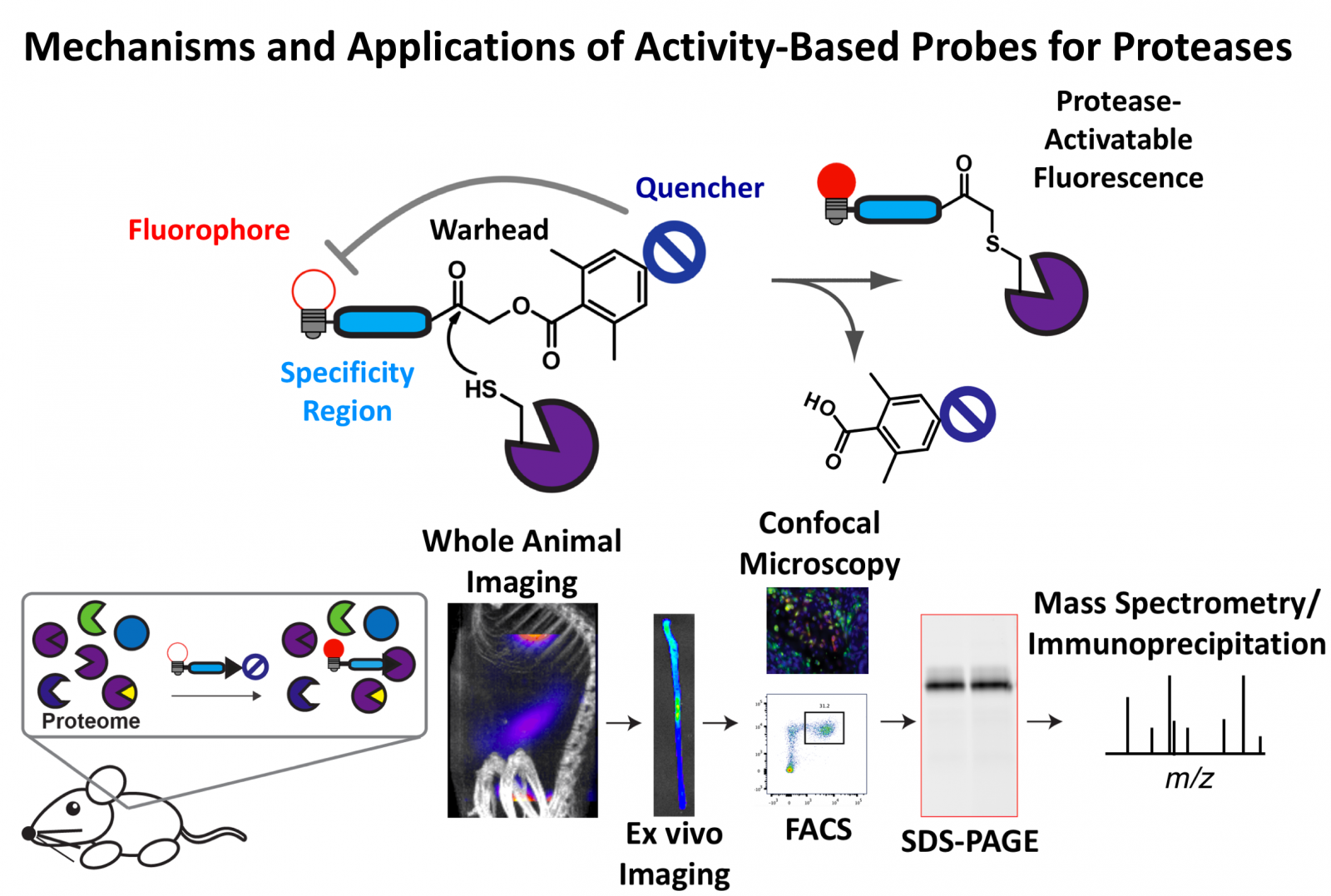
Proteases are enzymes that cleave peptide bonds of proteins. To prevent cleavage at the wrong place or time, and thus protect the body from aberrant proteolysis, most proteases are synthesised as inactive proteins called zymogens. They become activated in response to a conformational change, which can be mediated by alterations in pH or cleavage by other proteases. Once activated, proteases are also subject to spatial and temporal regulation by endogenous inhibitors. As a result of these complex modes of post-translational modification, traditional biochemical methods that survey total protein levels rarely reflect the pool of active, functional enzymes. The ability to specifically measure and modulate the activity of a protease in its native environment is therefore required to define its precise proteolytic functions during health and disease.
To achieve this, efforts from our team and others have focussed on developing activity-based probes (ABPs) for diverse cysteine and serine proteases. These tools capitalise on the catalytic mechanism of proteolysis, combining a protease recognition sequence with a reactive functional group called a warhead. We can apply these tools to measure protease activity by electrophoresis (in-gel fluorescence), confocal microscopy, flow cytometry, or by imaging whole tissues or even whole organisms. The covalent, irreversible nature of the probes allow us to identify their targets with absolute specificity through immunoprecipitation experiments or activity-based proteomics.
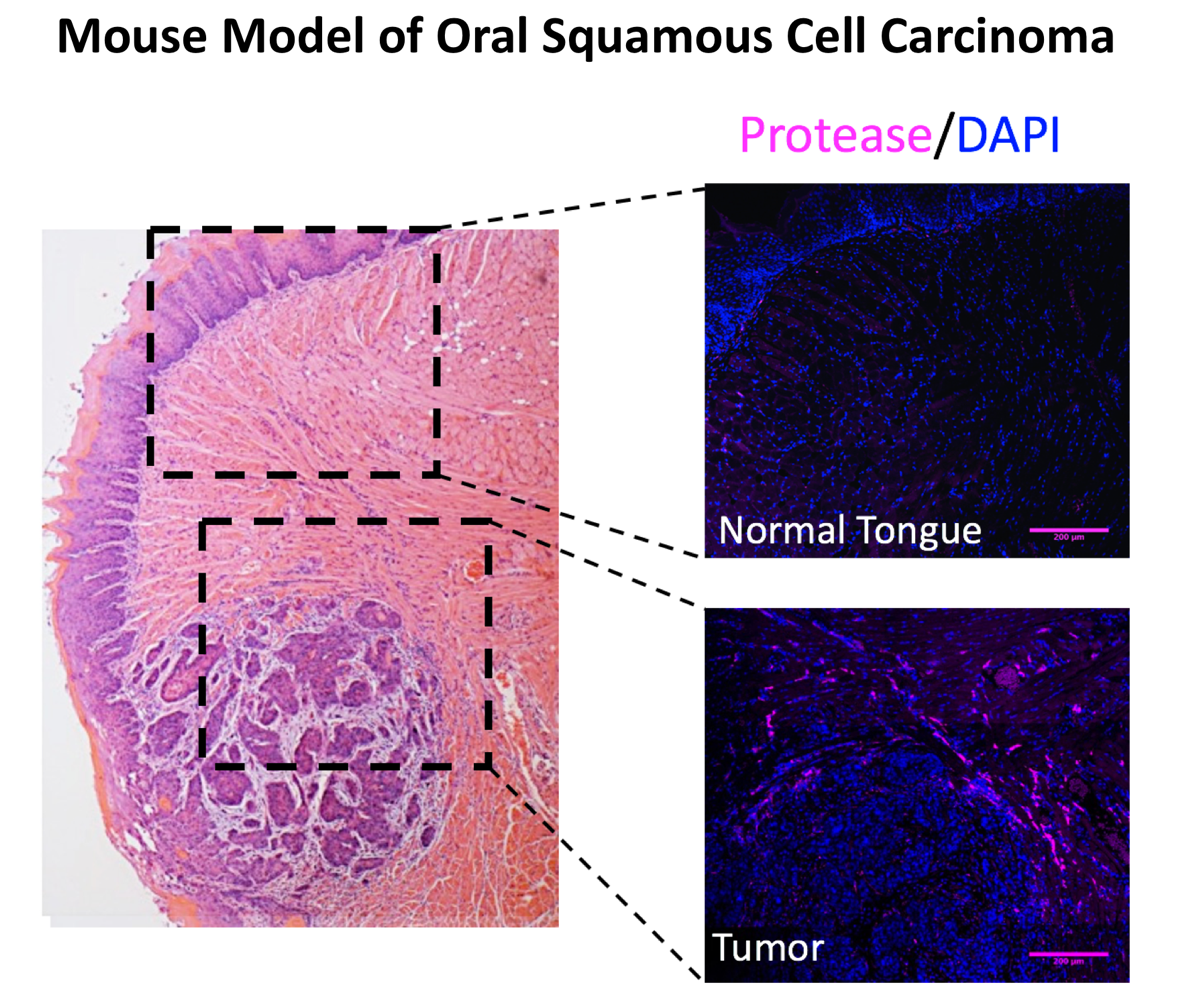
Understanding the contribution of proteases to oral cancer pain and metastasis
Oral squamous cell carcinoma is the most common head and neck cancer. Many patients with oral cancer experience severe pain that profoundly interferes with their quality of life. Treatments for this pain are currently limited to opioid drugs, which are not suitable for long-term use due to side effects such as sedation and constipation. Oral cancer often spreads to cervical lymph nodes, and once this metastasis occurs, patient survival rates drop below 40%. Current methods to predict the spread of oral cancer are ineffective; thus, most patients undergo radical elective neck dissection to remove all cervical lymph nodes prior to the appearance of metastatic lesions.
Our group is investigating the mechanisms by which proteases mediate oral cancer pain and metastasis. Our approach is multifaceted, and examines protease function in tumour and associated immune cells in vitro, in mouse models of oral cancer, and in clinical samples derived from oral cancer patients. We aim to establish proteases as biomarkers for diagnosing oral cancer, as predictive markers for the occurrence of cervical metastasis, and as drug targets for the treatment of oral cancer pain and prevention of metastasis.
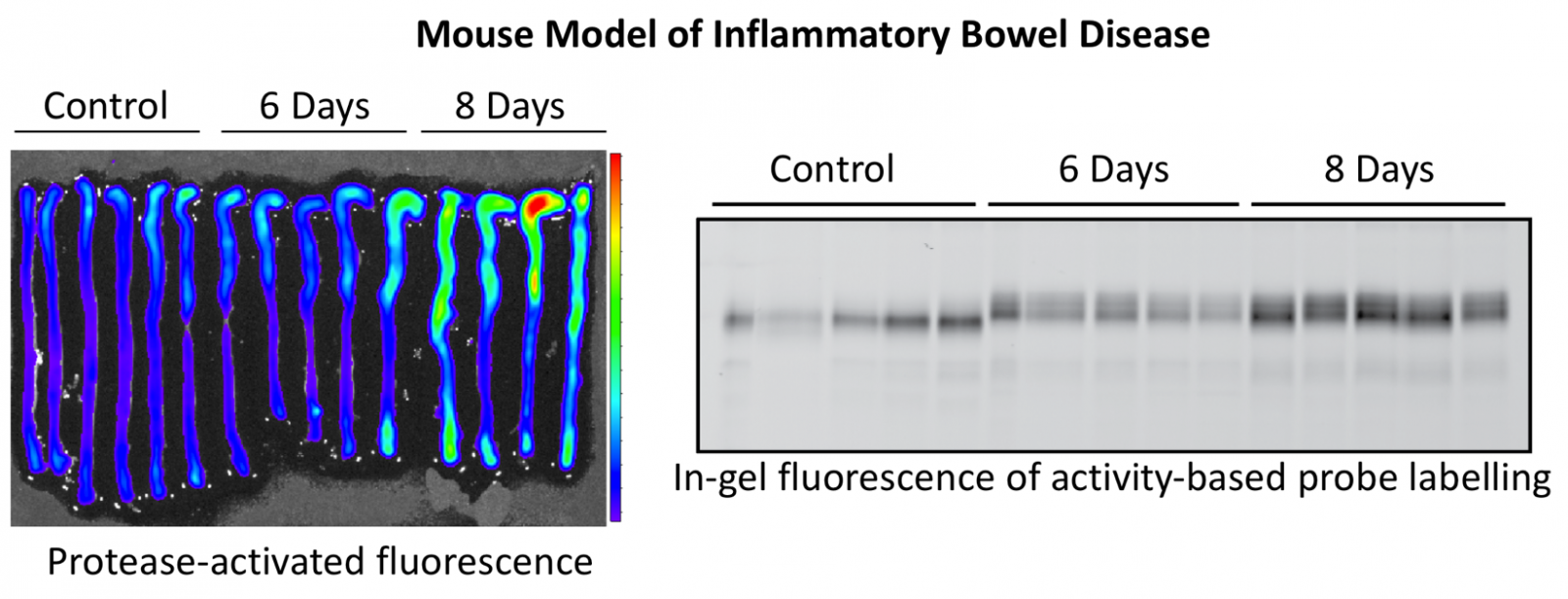
Proteases in inflammatory bowel disease
Using activity-based probes and protease inhibitors, we have identified a number of cysteine and serine proteases that are strongly upregulated in mouse models of ulcerative colitis and Crohn’s disease, as well as in mucosal biopsies from patients with these diseases. We seek to understand the function of these proteases, whether pro- or anti-inflammatory, with the goal of developing new diagnostic tests and therapeutics.
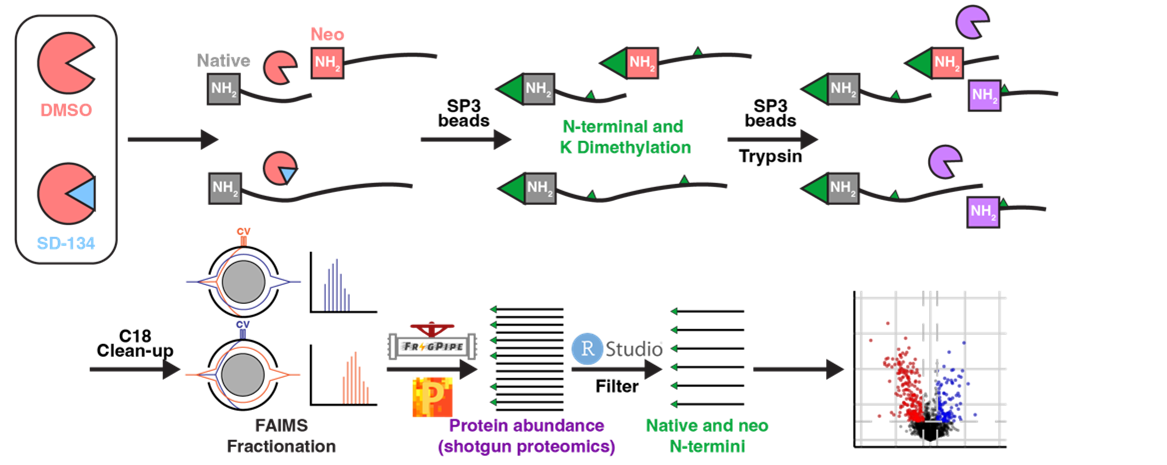
Application of proteomics to identify protease substrates in health and disease
Using innovative mass spectrometry-based proteomics methods, we can identify specific proteins that are cut by proteases. Knowledge of the substrates of a particular protease is essential for full understanding if its function and mechanisms of action. Knowing how this changes in the context of disease is critical for validating proteases as potential drug targets.
Techniques
- Protein biochemistry (electrophoresis, in-gel fluorescence, immunoblotting)
- Molecular biology (CRISPR gene editing)
- Confocal microscopy on cells and tissues
- Histology
- Activity-based proteomics and N-terminomics
- Animal models of disease (oral cancer, colitis)
- In vivo and ex vivo imaging
Extended Publication List on Google Scholar
Group Members
Research Fellow
Beth Anderson
PhD Students
Bangyan Xu
Lauren Bird (co-supervised with Hayley Newton)
Huw Morgan (co-supervised with Justine Mintern)
Mike Bozin (co-supervised with Nick Clemons)
Honours Students
Irene Lee
Piyapa Tantisirivat
Laboratory Head
Laura Edgington-Mitchell, Ph.D.
Senior Research Fellow
Department of Biochemistry and Pharmacology
School of Biomedical Sciences
Tel. (+61) 3 90354630
Laura.EdgingtonMitchell [at] unimelb.edu.au
Biography
Dr. Laura Edgington-Mitchell obtained her PhD in Cancer Biology from Stanford University in California in 2012. Studying under Prof. Matthew Bogyo, she specialised in synthesising activity-based probes (ABPs) to study the activity and function of proteases involved in cell death and cancer. She pioneered new methods to detect protease activity by noninvasive optical imaging. In 2013, Dr. Edgington-Mitchell initiated postdoctoral studies with Dr. Belinda Parker at La Trobe University. There, she used ABPs targeting cysteine cathepsin proteases to study their contribution during breast cancer metastasis to bone and revealed new functions for these enzymes during the differentiation of tumour-induced immune cells into osteoclasts. In 2015, she was awarded an NHMRC Peter Doherty Fellowship. Under the mentorship of Prof. Nigel Bunnett, she established an independent research program at the Monash Institute of Pharmaceutical Sciences. There, she continued to expand the repertoire of chemical tools to study diverse protease families and explored their roles in inflammatory bowel disease, irritable bowel syndrome, pancreatitis, and cancer. Her work on proteases in oral cancer in collaboration with Prof. Brian Schmidt led to an appointment as an Adjunct Associate Professor at the New York University College of Dentistry. In January 2018, Dr. Edgington-Mitchell was awarded a Grimwade Fellowship and an ARC DECRA Fellowship and joined the Department of Biochemistry and Molecular Biology, as a laboratory head. In 2022, she was reappointed as a Senior Lecturer in the Pharmacology Discipline. Building on a foundation in chemical biology, her research group aims to validate proteases as therapeutic targets in cancer and inflammation.