Hinde Group

“We are using fluorescence microscopy methods to visualise how molecules move throughout the 3D DNA network of a living cell. These analytical tools enable us to probe genome organization from the point of view of the proteins exploring this complex environment and recover the microstructure of the cell nucleus.” Elizabeth Hinde
Research
The research focus of the Hinde lab is on the architectural organisation of the cell nucleus and how chromatin dynamics facilitate navigation of the genome.
Inside the nucleus of a living cell, DNA is folded into a multi-layered 3D structure called chromatin. At any moment in time, thousands of proteins are diffusing throughout this structural framework - scanning the genome for a target DNA sequence. The question is “does the chromatin network serve as ‘road map’ for molecular traffic during DNA target search?” To investigate this, we are developing microscopy methods - based on fluorescence lifetime and correlation spectroscopy - to track how proteins move throughout the DNA networks of a living cell. Using this technology we have discovered sub-micron rearrangements in chromatin density that direct the diffusive route of DNA repair factors to damage sites and mediate transcription factor accumulation at specific nuclear locations. This body of work demonstrates an active role for 3D chromatin organisation in nuclear factor navigation. This is important because a hallmark of cancer is genome dysregulation. We therefore now aim to investigate how disruptions to chromatin structure redirect nuclear traffic and the implications this has for maintaining genome function. The current projects in the lab are:
1. Mapping the spatial connectivity of nuclear architecture in health and disease.
2. Chromatin dynamics during the DNA damage response.
3. The role of protein oligomerisation in transcription factor DNA target search.
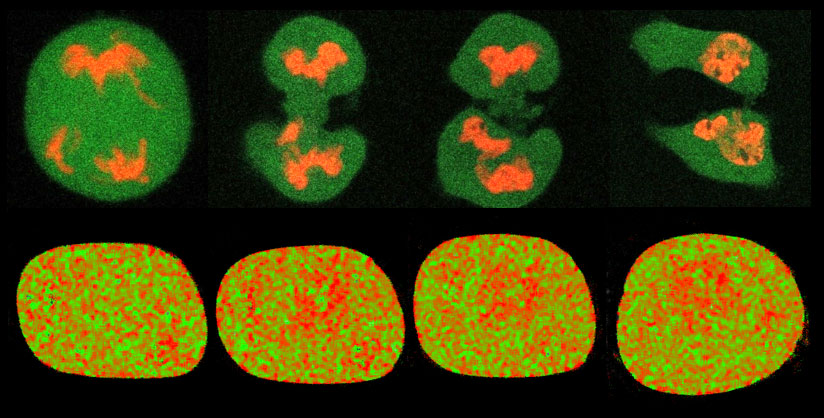
Figure 1. Chromatin dynamics range from global reorganisation of the genome during the cell cycle on a minute-to-hour timescale (top row) down to nanometer-to-micron rearrangements in local chromatin compaction during the DNA damage response (bottom row).
Methods
Advanced fluorescence microscopy has revealed molecular insights into the spatial dynamics of biological processes that could not have been obtained by any other means. In the Hinde lab we are applying biophysical methods of analysis based on fluorescence lifetime imaging microscopy (FLIM) and fluorescence fluctuation spectroscopy (FFS) to quantitate protein interaction, aggregation, binding kinetics and diffusion in live cells. For example we use:
- Fluorescence lifetime imaging microscopy (FLIM) – FRET biosensor detection
- Fluorescence correlation spectroscopy (FCS) – protein diffusion and concentration
- Spatiotemporal correlation spectroscopy (STICS, RICS) - protein binding dynamics
- Pair correlation analysis (pCF, pCOMB) – spatial route of protein diffusion
- Moment based brightness analysis (NB, PCH) – protein oligomerisation
A particular interest of the lab is the development of microscopy methods based on spatial pair correlation function (pCF) analysis to extract the molecular connectivity of nuclear architecture with sub-micron resolution. We have demonstrated that by combining pair correlation analysis with fluorescence lifetime and brightness information we can track nuclear factor complex formation within the structural framework of the nucleus.
Group Members
Group Leader
Elizabeth Hinde
Postdoctoral Scientist
Jieqiong Lou
Adele Kerjouan
PhD Student
Ashleigh Solano
Masters Student
Zhen (Jen) Liang
Alex Hopper
Biography
 Contact:
Contact:
Email: Elizabeth.hinde[@]unimelb.edu.au
Tel: (03) 8344 9188
Elizabeth Hinde is currently the Jacob Haimson and Beverly Mecklenburg Lecturer in the School of Physics, University of Melbourne, and holds a joint appointment with the Department of Biochemistry and Molecular Biology as an NHMRC Career Development Fellow. In 2010 Elizabeth completed her PhD in fluorescence spectroscopy at the University of Melbourne and was then recruited to the University of California, Irvine, USA to pursue a post-doctoral fellowship under the mentorship of Professor Enrico Gratton. In the Gratton lab (2010-2013) Elizabeth developed methods based on fluorescence correlation spectroscopy (FCS) and fluorescence lifetime imaging microscopy (FLIM) to quantify chromatin dynamics in live cells. With the aim of applying this technology to cell biology, Elizabeth returned to Australia as a UNSW Vice Chancellor Fellow (2013-2015) and a Cancer Institute NSW Early Career Fellow (2015-2016) within the EMBL Australia node for Single Molecule Science, University of New South Wales (UNSW). Under the mentorship of Prof. Katharina Gaus, Elizabeth established a research program at UNSW which investigated live cell nuclear organisation. Collectively Elizabeth's work has been recognised by the US Biophysical Society with the 2014 Young Fluorescence Investigator Award and the Australian Society of Biophysics with the 2016 McAulay-Hope Prize for Original Biophysics.

