Ghosal Group

"We are harnessing this unique power of cryo-ET and combining it with correlative light and electron microscopy (CLEM), and Focused Ion Beam (FIB) milling to elucidate the structure and function of large bacterial macromolecular assemblies in their native context inside ‘living’ cells." Debnath Ghosal
Contact
Email: debnath.ghosal [at] unimelb.edu.au
Biography
Dr Debnath Ghosal is a Senior Lecturer and laboratory head in the Department of Biochemistry and Molecular Biology, University of Melbourne at the Bio21 Molecular Science and Biotechnology Institute. His research interest lies in the intricacies of host-pathogen interaction particularly, how bacterial and viral pathogens utilize complex molecular machines to cause infection.
Debnath obtained his Master's degree from the Tata Institute of Fundamental Research (Mumbai, India) and subsequently completed his PhD dissertation from the MRC Laboratory of Molecular Biology (University of Cambridge, UK) under the supervision of Dr Jan Löwe (FRS). After his PhD, Debnath worked briefly as a career development fellow at the MRC LMB before moving to California Institute of Technology (USA) for his postdoctoral training with Prof Grant Jensen. He has more than ten years of research experience in the areas of structural biology, biochemistry, and bacterial cell biology.
Debnath joined University of Melbourne in early 2020. His laboratory utilizes the unique power of cryo-ET and combines it with correlative light and electron microscopy (CLEM), and Focused Ion Beam (FIB) milling to investigate the structure and function of large bacterial macromolecular assemblies and host-pathogen interaction.
More about Ghosal laboratory here: www.ghosallab.com
Research
Bacterial cells were long thought to be “bags of enzymes” with little or no intracellular organization. However, over the last three decades, the advent of fluorescent proteins and development of new imaging modalities (e.g. electron cryotomography, super-resolution light microscopy etc.) have revolutionized our understanding of the bacterial domain of life. To survive efficiently in its environment, the bacterial cell requires a high spatiotemporal organization both structurally and metabolically. They utilize sophisticated sensory and regulation pathways to orchestrate motility and cell-cell communication, assemble complex molecular machines to inject effectors into neighbouring bacterial/eukaryotic cells, amass nutrients inside specialized compartments, possess eukaryotic homologues of actin, tubulin and intermediate filaments, and even some of them own membrane bound organelles and distinct cellular compartmentalization. This paradigm shift in our understanding has opened up new opportunities to explore ever increasing internal complexities within bacterial cells!
Electron cryotomography (cryo-ET) has unrivalled power to visualize the native structure of macromolecular complexes in their mise-en-scène, i.e., inside ‘intact’ cells and thus provides unprecedented mechanistic and structural insights. In this method, plunge-frozen vitreous samples are iteratively imaged in a transmission electron microscope (TEM) as they are tilted, resulting in a stack of 2D images (tilt-series) that can be reconstructed into a 3D volume or a “tomogram”. With resolutions of ~2-5 nm, cryo-ET uniquely bridges the “resolution gap” between atomic structures and gross cellular morphology. Additionally, recent improvements in software, hardware and the implementation of improved subvolume averaging methods have allowed cryo-ET to enter the realm of subnanometer resolution. In addition to structural information, cryo-ET also provides pivotal insights into the distribution pattern of protein complexes in cells, their relative orientations and structural dynamics making this an ideal method for ‘visual proteomics’ and in-cell structural biology. We are harnessing this unique power of cryo-ET and combining it with correlative light and electron microscopy (CLEM), and Focused Ion Beam (FIB) milling to elucidate the structure and function of large bacterial macromolecular assemblies in their native context inside ‘living’ cells.
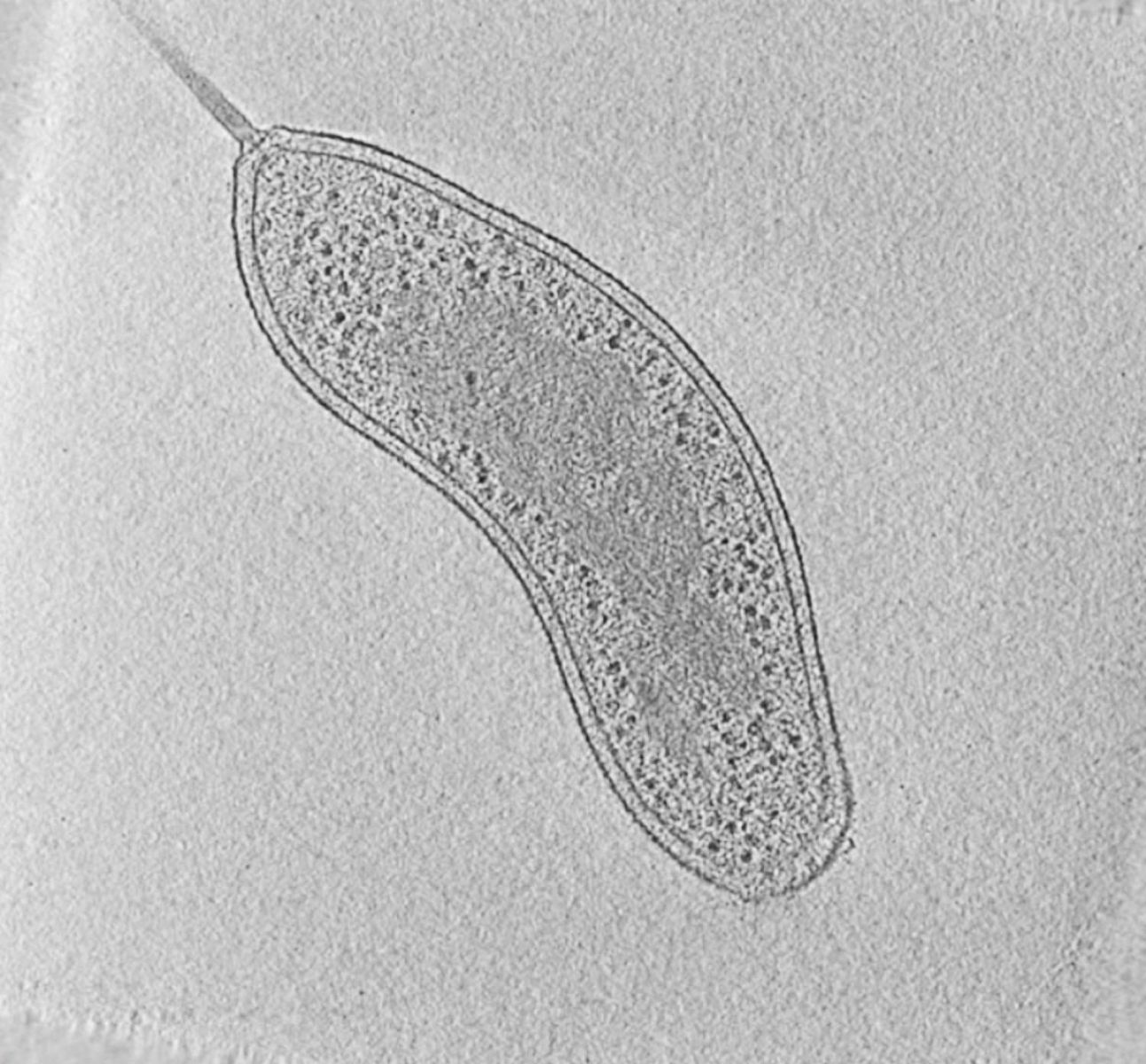 |
Fig. 1: Cryotomogram of an intact Bdellovibrio bacteriovorus cell. Visible cellular structures are segmented and annotated. Left: tomographic slice, right: segmented tomogram. Courtesy of Grant Jensen lab.
We complement cryo-ET with other biochemical and biophysical techniques such as in vitro biochemical reconstitution, genetic and functional analysis, fluorescence imaging and high-resolution structural techniques such as X-ray crystallography and cryo-EM.
Research projects:
We are particularly interested in bacterial molecular machines that are responsible for toxin delivery (bacterial secretion systems), and bacterial cytoskeletal (rather, bacterial ‘cytomotive’) filaments that execute a diverse array of cellular functions.
1. Bacterial Secretion systems
2. Structure and function of bacterial ‘cytomotive’ filaments


