'Humpty Dumpty' program 'Bandage' helps piece DNA sequences back together again and wins 2015 iAwards
During his Masters of Bioinformatics degree, Ryan Wick was studying antibiotic resistance in bacteria, such as Salmonella.
 He set about analysing the bacterial genome – all of its DNA - in order to find the antibiotic-resistant mutations that give it a survival advantage.
He set about analysing the bacterial genome – all of its DNA - in order to find the antibiotic-resistant mutations that give it a survival advantage.
The process of DNA sequencing chops up bacterial DNA into millions of small fragments.
Like the remains of Humpty Dumpty, the challenge is then to put all the fragments back together again.
Ryan used genome assembler programs which generate pages of DNA letters: Ts and As and Gs, and Cs, representing the nucleotide bases.
But what did it all mean? How did they fit together and where should he start looking for the antibiotic resistant genes? These reams of letters were not ‘human’ readable!
Ryan enquired amongst his colleagues in the lab about which programs they used to analyse the data, called an ‘assembly graph’.
“I assumed there would be a program,” explained Ryan. “There was nothing out there.”
So, he set about creating the program that he needed. “I wanted to make something I could use,” he explains.
‘Bandage’ (a Bioinformatics-Application-for-Navigating-De-novo- -Assembly-Graphs-Easily) is a program that makes it possible to put the pieces back together again.
For his invention, Ryan Wick, MSc (Bioinformatics) student with the Holt group, was named the 2015 National Winner at the iAwards, 27 August. The iAwards recognise Australian achievements in technology innovations and are judged by the ICT industry.
The Victorian Life Sciences Computation Initiative (VLSCI) reports that the major value of the application is that it “visualises genome assembly graphs in an interactive way, providing insights into genetic data that were not previously possible, with potential to provide significant benefits to the health sector.”
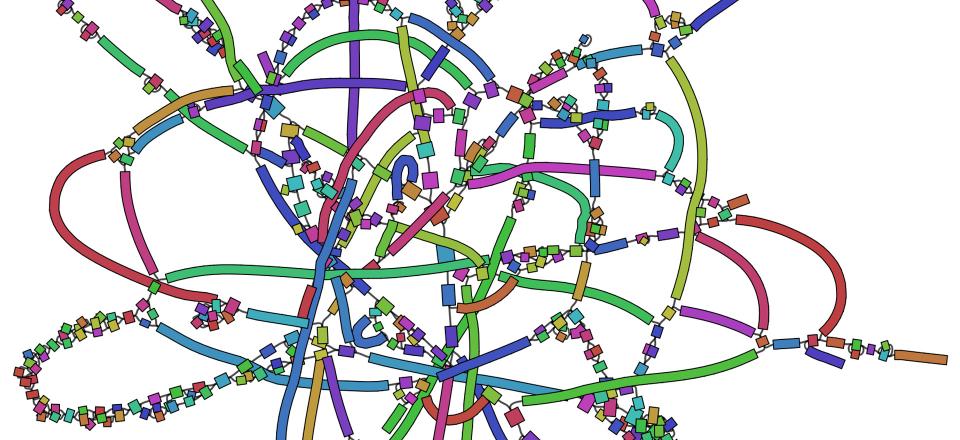
‘Bandage’ represents the assembled DNA fragments, of varying length, in a visual way and shows which sequences potentially connect to which others.
Bacterial DNA in its original form occurs in a circular chromosome. Due to the many repeats of small fragments it is not possible to reassemble a perfect circle in the exact order that it occurred in the organism, explains Ryan. However, Bandage is useful for visualising what fragments may connect to each other. It’s then up to the researcher to ‘detangle’ the fragments.
“The image created by Bandage sometimes looks a bit like a tangle of rubber bands, hence the name “Bandage”,” explains Ryan.
.png)
Interestingly, it’s in these tangles or knots of small repeated nucleotide fragments, where antibiotic resistance genes are commonly found.
“It’s a starting point that helps you know where to look,” explains Ryan. Also, there is a certain element of reproducibility of the images for sequenced genomes of particular species.
Whether it is to discover antibiotic resistant genes in bacteria, or to find unique mutations in cancer cells that cause them to metastasize, the ‘Bandage’ program provides a means of visualising genomic data, to make it possible to observe patterns and to understand how genetic sequences fit together and influence each other.
Bandage is Open Source software, that can be used and downloaded free of charge. “I am excited that researchers are already using and modifying the software for their own unique situations,” said Ryan. “I actually felt a bit out of place at the iAwards,” adds Ryan -
“this invention is free and I did not set out to make a profit.”
To learn more about BANDAGE and to download the program, follow this link. [https://rrwick.github.io/Bandage/]
Ryan developed Bandage under the guidance of VLSCI’s Acting Director, Professor Justin Zobel; VLSCI power user and Bio21 Group Leader, Dr Kat Holt; and Dr Mark Schultz from the Holt lab.
By Florienne Loder and Maggie Scott

