From decades to days: 3D modelling tuberculosis
Associate Professor Justin Denholm, Dr David Ascher and Malancha Karmakar
While many of us may think of tuberculosis (TB) as a historical disease, it actually remains one of the top ten causes of death worldwide. While it’s relatively uncommon in Australia, in 2016, around 10.4 million people fell ill with TB globally and 1.7 million lost their lives as a result of the disease.
Part of the reason that TB remains such a difficult disease to manage is the long time it can take to establish what treatment is best for each patient. Traditional approaches to demonstrating that a specific medication will be effective against a strain of TB involve laboratories culturing the organism and checking whether the drug stops its growth.
And because TB is very slow growing, this process can take weeks or even months; and that is long enough for resistance to develop. The end result is that those suffering may die before the right treatment can be started.
In recent years, genomics has brought advances to TB diagnosis, including the ability to rapidly identify when strains have mutations likely to cause drug resistance.
Tuberculosis: Still a major killer
Even so, when a new mutation is found, understanding whether it can lead to drug resistance means seeing evidence both in laboratory testing and in clinical settings. Clinicians and TB programs, then, may need to treat people while there’s still uncertainty about the meaning of these mutations.
But now innovative technology is providing us with a game changer for treatment – modelling mutating tuberculosis genes in a matter of hours.
Modelling mutations
Our team has developed a computer-generated model that allows clinicians to tailor effective therapies for individual patients with multidrug-resistant tuberculosis (MDR-TB).
Researchers from The Peter Doherty Institute for Infection and Immunity and University of Melbourne’s Bio21 Molecular Science & Biotechnology Institute, led by University of Melbourne PhD student Malancha Karmakar, have devised a 3D-computational approach to predicting the impact of mutations in TB.
This work is aimed at supporting global programs that are starting to use new TB therapies, allowing them to be tailored to an individual as early as possible while also avoiding the use of ineffective and harmful treatments.

While our research was underway, doctors in Melbourne were treating a patient with multidrug-resistant (MDR) TB who was experiencing significant side effects from the cocktail of medications they required. These problems with side effects are sadly common, as MDR-TB needs treatment for up to two years with medications that can themselves cause deafness, liver and thyroid damage, joint pain and other issues that add to the difficulty of completing therapy.
Scientists at the Victorian Infectious Diseases Reference Laboratory used this cutting-edge genome sequencing technology to look for drug-resistance mutations to help tailor the patient’s therapy and identified a mutation that had never been reported before.
The chemical warfare against bacterial superbugs
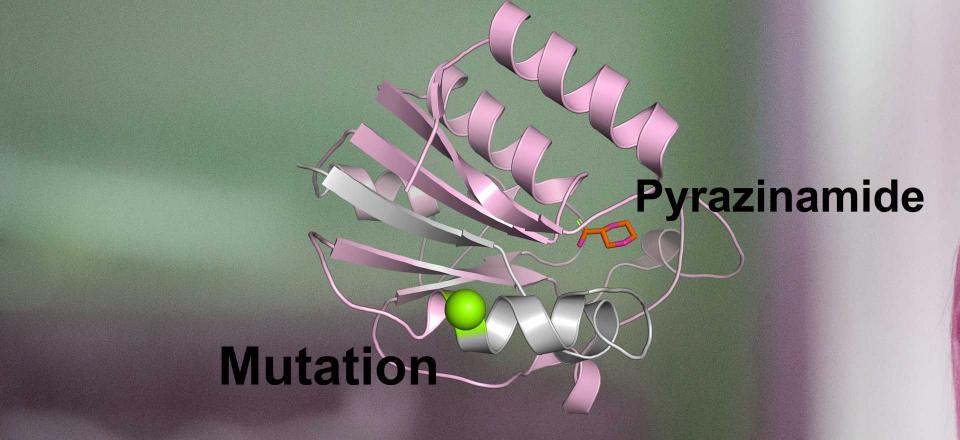
Suspecting that the mutation might be causing resistance, Ms Karmakar was quickly able to build a new 3D model to investigate. What she found was that the mutation made one of the drugs being used completely ineffective. As a result, doctors swapped it for another, more effective drug.
From decades to days
What this shows us is that this new tool is a game-changer. What would normally take decades could now just take a matter of hours.
Right now, in order to use genetic tests to reliably predict whether a drug will be effective, a person needs to have been treated with a drug, have that therapy fail and then be found to have a gene mutation.
And it doesn’t stop there. This is followed up by identifying enough people around the world that have that pattern to inform what treatment you prescribe for that particular mutation. It can take years.
By understanding how mutations work within 3D space, the team can identify likely resistant mutations that have never arisen before.
This is a huge boost to drug-resistant TB programs, which globally have to take a ‘best guess’ approach, where they give the same, standardised therapy to people with MDRTB, with little capacity to adjust in real-time.
And the research brings together researchers from across the discipline – clinicians, laboratory scientists, bioinformatics experts and computer scientists all helped identify this mutation and its significance.
What this means is that the modelling can be done quickly, making results available in ‘real time’. This truly is a ‘bedside to bench and back’ approach, that we’re now planning to expand to a range of other genes that may cause TB drug resistance.
More importantly, once the tool is fully developed it will be available on the web, meaning clinicians around the world can enter the resistance mutations into the system – helping to create and build effective treatments for their patients.
Banner Image: Shutterstock
“This article was first published on Pursuit. Read the original article.”